Physical Society Colloquium
Mining the molecular noise: fluorescence fluctuation
image analysis reveals protein interactions and transport in living cells
Departments of Physics and Chemistry
McGill University
I will start with an overview of fluctuation analysis from statistical
physics and that provides the basis for fluorescence correlation
spectroscopy (FCS) analysis that was originally developed to study dye
binding kinetics and protein transport properties in cells.
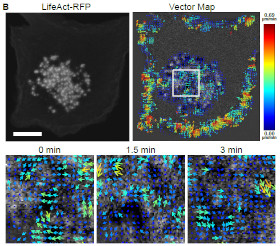
Actin waves revealed but STICS in a podosome cluster in immune dendritic cells
The transport properties of biomolecules in cells can reveal a great deal
about the functional interactions regulating cells at the molecular level.
Various biophysical methods have been developed to measure these properties
in cells, although most have relied on fluorescence microscopy imaging as
the window for measurement of labeled macromolecules in living cells. Image
correlation methods are an extension of fluorescence correlation
spectroscopy that can measure protein-protein interactions and
macromolecular transport properties from input fluorescence microscopy
images of living cells. These approaches are based on space and time
correlation analysis of fluctuations in fluorescence intensity within
images recorded as a time series using a fluorescence or super-resolution
microscope. I will introduce spatio-temporal image correlation spectroscopy
(STICS) and its 2 color cross-correlation variant (STICCS) and show how the
analysis can reveal hidden coupling between retrograde cellular actin flows
and the plasma membrane lipids for activated Jurkat cells. I will then
describe the application of the STICS and pair vector correlation for
measuring cellular waves of adhesion related macromolecules talin and
vinculin as well as cytoskeletal actin between assembling and disassembling
podosomes in dendritic immune cells. Podosomes are cylindrical membrane
complexes with an integrin adhesive ring and an actin rich core that are
associated with cellular migration and invasion in specific cell types. EM
and super-resolution microscopy of cells shows radial actin filaments that
connect neighboring podosomes. The image correlation analysis combined with
pharmacological perturbation experiments show that podosome turnover is
coordinated within local clusters in cells with a correlation length scale
extending to next nearest neighbor podosomes. I will highlight, our recent
work pairing radial STICS with live cell super-resolution microscopy
reveals that the dynamic coordination between podosomes differs for cells
on soft versus stiff substrates which provides clues to the mechanistic
function of podosomes. If there is sufficient time I will cover recent
applications of k-space image correlation spectroscopy to simultaneously
measure fluorescent protein diffusion and fluorescent probe emission
blinking kinetics in living cells.
Friday, October 22nd 2021, 15:30
Ernest Rutherford Physics Building, Keys Auditorium (room 112)
Colloquium recording
|