|
Biological PhysicsMcGill Physics is growing a strong and highly collaborative biophysics research community, including 5 in-Department and several out-of-Department members. Their active research programs are seeking highly motivated graduate students and researchers.
 Velocity map... »
Velocity map of retrograde transport of alpha-actinin/EGFP in a mouse
fibroblast cell. Measured by STICS analysis.
 DNA and beads... »
Bottom Left: A cartoon depicting a bead optically trapped inside a
nanochannel with an extended DNA molecule (shown in red). The DNA molecule
is driven against the bead at a fixed sliding speed V. Top: Time-series of
fluorescence micrographs showing a YOYO-1 stained T4 DNA molecule ramming
against a bead. Note that the DNA molecule undergoes transient dynamics and
compresses against the bead, leading to a decrease in molecule extension
and increase in chain concentration along the channel. Bottom Right:
DNA compression as a function of sliding speed.
 Multi-colour fluorescence excitation systems...»
Multi-colour excitation systems use lasers of different wavelengths to
simultaneously image different molecular species. By tracking the dynamics
and interactions of multi-component biomolecular systems in real time,
the mechanisms underlying molecular dances can be newly visualized and
understood. New microscopy technologies and analysis methods that extend
the range of accessible interaction timescales and affinities enable
unprecedented understanding of biophysical systems and push the limits
of biomolecular diagnostics.
Visualizing dynamics and interactions between biomolecules (e.g. protein, DNA) with single-molecule resolution allows for the biophysical mechanisms underlying life preserving processes such as DNA transcription and repair to be newly uncovered and understood (Leslie; Brouhard, Biology, associate member in Physics). Furthermore, combining nanoscale imaging devices, wide field image correlation spectroscopy methodologies and DNA nanostructures, in a collaborative effort between Leslie, Wiseman and collaborators in the Department of Chemistry, opens the door to creating ultra-sensitive biomedical diagnostics (e.g. of biomarkers that indicate cancer onset). As well there are collaborations that involve studies in fields related to neuroscience, such as synaptic formation and neuron migration (Grütter and Wiseman; Neurophysics Training Grant Program and Neuroengineering); and to mechanosensing, such as sensors to measure molecular motor systems (Grütter and collaborators at the Meakins Christie Chest Hospital and the Bone Centre; Rassier, Kinesiology, associate member in Physics), or to developmental biology such as embryonic growth and patterning (François and Pourquié group at the IGBMC Strasbourg, France). 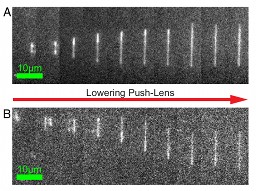 Manipulating and trapping DNA with CLINT ...»
Convex lens-induced nanoscale templating (CLINT) allows us to dynamically
manipulate and trap single DNA molecules. In the above frames, a single
λ-DNA molecule extends in a nano-channel while the push-lens is
lowered. (A) Extension in a 27-nm channel. The time between frames is 364
ms. (B) Extension in a 50-nm channel. The time between frames is 910 ms.
Students and postdoctoral researchers trained in this environment gain quantitative and interdisciplinary skills, and can come from biological or physics backgrounds. Our biophysics research programs offer students and postdoctoral researchers the opportunity to gain expertise in state-of-the-art two-photon and nonlinear microscopy, image correlation spectroscopy and other fluctuation-based methods, direct tracking in live cell methods, confocal microscopy, computational biology, lasers and optical trapping, atomic force microscopy, protein-engineering, signal transduction, gene expression, neurophysiology, micro/nanofluidic bioanalysis device fabrication, nanoparticle labels, and total internal refection and fluorescence resonance energy transfer microscopies. [1] Bruce Alberts, Molecular Biology of the Cell, 5th Edition (Taylor & Francis, 2007). 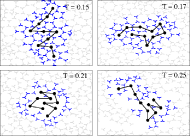 Cold protein ...»
Characteristic configurations of a protein in cold water (T=0.15 and
T=0.17), at an intermediate temperature (T=0.21), and in hot water
(T=0.25). The distance of highlighted (shell) water molecules to the
protein is less than 2.5 in units of Rh. In cold water, the monomers
are typically surrounded by clathrate-like cages.
 Bacterial spheroplast »
Bacterial spheroplast with its mechanosensitive channel protein
fluorescently-labeled, in preparation for insertion into a supported
planar lipid bilayer for direct study by fluorescence microscopy of the
distribution, dynamics, and possible tension-induced cooperative gating
of the stretch channels.
|